Carola Wenk's web pages.
![[LOGO]](./spots2.jpg) |
Analysis of 2D
Electrophoresis Gels
|
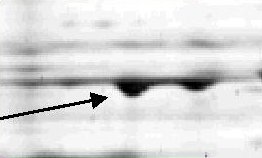
In proteomics, which is the field of protein analysis, protein samples
are separated using a technology called two-dimensional gel
electrophoresis (2-DE). This
produces gel images
in which every dark spot
represents one protein. Eventually one would like to compare several
images (which is basically a point matching problem; it is similar to
considering
the starry sky and comparing different star constellations), which is
called gel matching. But first one needs to segment the image into
the spot areas (spot detection). Both tasks are not
straight-forward, and still need a lot of human help nowadays, which is
slow and costly.
Pharmaceutical companies are very much interested in automating these
processes as much as possible.
Spot detection and spot matching across multiple sets of gel images
are essential first steps for proteomics investigations based on
two-dimensional gel electrophoresis. There are a number of
software packages on the market that can be used for this purpose,
with each offering distinct advantages and disadvantages. A major
difference between many of these programs are the algorithms used for
gel matching and spot detection. A high-quality spot detection
algorithm is fundamental since accurately detected spots are the basis
for all further analyses. A high-quality gel matching algorithm is
essential in order to minimize manual postprocessing and in order to
facilitate fast cross-matching between sets of gels. However at this time,
there is no consensus about which program/algorithm provides the most
accurate assessment of changes in protein levels among sample groups.
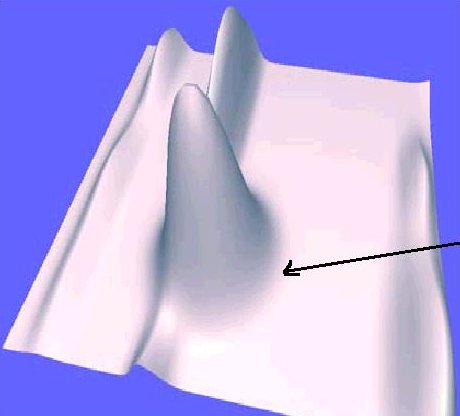
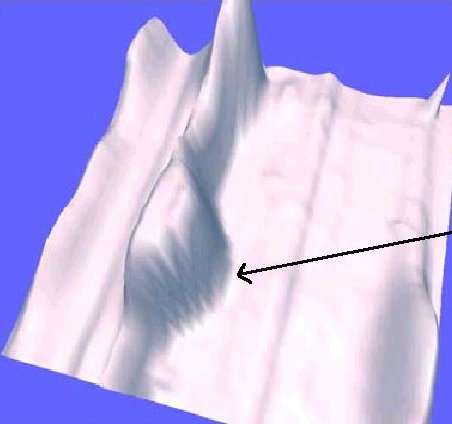
A 3-dimensional Gaussian distribution (see left) is a widely accepted modeling
assumption for protein spots, which is used in several spot detection
algorithms. It is however clear from the 2-D gels (see right) obtained from Sue
Weintraub's laboratory at the UT Health Science Center at San Antonio
that many spots exhibit reproducible intensity patterns that do not
fit a Gaussian model.
This project investigates new spot detection and gel matching
algorithms. The CAROL software [Hoffmann et al. 1999,
Pleissner et al. 1999, Kriegel
et al. 2000] has been integrated into the gel analysis software
package PDQuest by Bio-Rad. Our current research focusses on new spot
models, new spot detection algorithms, faster gel matching algorithms,
and the integration of gel matching data with microarray data.
Current collaborators:
Former Students:
- Drew Shaw (undergraduate student)
- Sean Pivek (undergraduate student)
Publications:
- ``Models for Teardrop Spots in 2-DE Gels'' [abstract, poster] (S.M. Pivek, S.T.
Weintraub, and C. Wenk),
poster, 11th RCMI International Symposium on Health Disparities, 2008.
- ``Covering with Ellipses''
[ps.gz, pdf]
(A. Efrat, F. Hoffmann, C. Knauer, K. Kriegel, G. Rote, and C. Wenk),
Algorithmica (special issue on
shape algorithmics) 38(2):145-160, 2004.
- ``Covering Shapes by Ellipses''
[ps.gz, pdf]
(A. Efrat, F. Hoffmann, C. Knauer, K. Kriegel, G. Rote, and C. Wenk),
Proc. 13th Symp. Discrete Algorithms (SODA): 453-454, 2002, San
Francisco, USA
- ``Geometric Algorithms for the Analysis of 2D-Electrophoresis Gels'' [ps.gz, pdf]
(A. Efrat, F. Hoffmann, K. Kriegel, C. Schultz, and C. Wenk),
Journal of Computational Biology; special issue dedicated to RECOMB 2001,
9(2): 299-316, 2002.
- CAROL - a tool for spot detection in and matching of
two-dimensional electrophoresis gels: http://gelmatching.inf.fu-berlin.de (K. Kriegel, C. Wenk,
C. Schultz, F. Hoffmann, and D. Dimitrov). Since 2001 integrated into
the gel analysis software PDQUEST by BioRad.
- ``Geometric Algorithms for the Analysis of
2D-Electrophoresis Gels'' [ps.gz, pdf] (A. Efrat, F. Hoffmann, K. Kriegel,
C. Schultz, and C. Wenk), Proc. 5th Ann. Int. Conf. Computational
Molecular Biology (RECOMB): 114-123, 2001, Montreal, Canada
- ``An alternative approach to
deal with geometric uncertainties in computer analysis of two-dimensional
electrophoresis gels'' (K. Kriegel, I. Seefeldt, F. Hoffmann, C. Schultz,
C. Wenk, V. Regitz-Zagrosek, H. Oswald, and E. Fleck),
Electrophoresis 21:2637-2640, 2000
- ``New algorithmic approaches to protein spot detection and
pattern matching in two-dimensional electrophoresis gel databases''
(K.-P. Pleißner, F. Hoffmann, K. Kriegel, C. Wenk, S. Wegner,
A. Sahlströhm, H. Oswald, H. Alt, and E. Fleck), Electrophoresis
20:755-765, 1999
- ``An applied point pattern matching
problem: comparing 2D patterns of protein spots'' [ps.gz, pdf] (F. Hoffmann, K.
Kriegel, and C. Wenk), Discrete Applied Mathematics 3: 75-88,
1999
- ``Matching 2D patterns of protein
spots'' [ps.gz, pdf] (F. Hoffmann, K. Kriegel, and C. Wenk), Proc. 14th
Ann. Symp. Computational Geometry (SoCG): 231-239, 1998,
Minneapolis, USA
- ``Identification of Proteins by
Point Pattern Matching of Two-Dimensional Gel Electrophoresis Databases''
(H. Alt, F. Hoffmann, K. Kriegel, C. Wenk, E. Fleck, H. Oswald,
K.-P. Pleissner, S. Wegener), Jahrestagung der Humangenetischen
Gesellschaft, October 1998
- ``CAROL - New Algorithmic Tools for Comparing Two-Dimensional
Electrophoretic Gel Images'' [ps.gz, pdf] (H. Alt, F. Hoffmann, K. Kriegel,
C. Wenk, and K.-P. Pleissner), Poster P21, Electrophorese
Forum 1997, Strasbourg
Grants:
- ``Developing New Spot
Detection Algorithms for 2-Dimensional Gel Electrophoresis Image
Analysis'', collaboration with Sue Weintraub from UTHSCSA, subgrant of NIH RCMI Center Grant with PI James Bower, 9/2006 - 2/2007,
$10,000
- "CAREER: Application and Theory of Geometric Shape
Handling", NSF CCF-0643597, $400,468 , 3/1/2007 - 2/29/2012.
Last modified by Carola Wenk,
cwenk -at- tulane -dot- edu ,
![[LOGO]](./spots2.jpg)
![[LOGO]](./spots2.jpg)